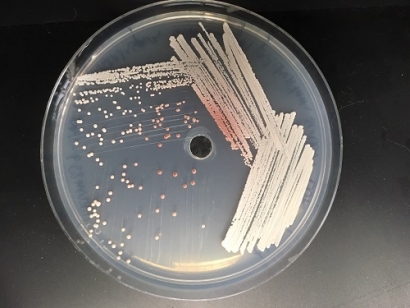
The pigment is synthesized in part through the same biochemical pathway researchers hope to use to improve production of isobutanol, a promising biofuel alternative to ethanol. In research published this week in the Proceedings of the National Academy of Sciences, a team based at the University of Wisconsin–Madison and the Great Lakes Bioenergy Research Center describe the genetic machinery that yeasts use to make pulcherrimin, which binds iron, an essential nutrient. The work is a key step toward harnessing the synthesis pathway for large-scale production of isobutanol as a biofuel.
“Compared to first-generation biofuels, such as ethanol, isobutanol has a higher energy content, blends better with gasoline, causes less corrosion, and is more compatible with existing engine technology,” said GLBRC researcher Chris Todd Hittinger, a UW–Madison genetics professor who led the research.
Yeasts typically do not produce much isobutanol under normal conditions, explained David Krause, a postdoctoral fellow with GLBRC and the lead author of the new study. Most commonly studied species produce ethanol during fermentation. The early steps of isobutanol synthesis are the same as those used to make pulcherrimin pigment, and this caught the researchers’ attention.
“Our thought is that these yeasts that are making pulcherrimin may be primed in a way to make more isobutanol,” said Krause.
The researchers used comparative genomics spanning 90 yeast species to identify the genes involved in pulcherrimin production. They found a cluster of four genes, which they named PUL1-4, that seem to play complementary roles. Through extensive genetic characterization, they determined that PUL1 and PUL2 are required to make the molecule, while PUL3 and PUL4 appear to help the yeast transport it and regulate its production.
The discovery was surprising in part because it marks the first report of a gene cluster in budding yeasts responsible for producing a type of compound known as a secondary metabolite. Many secondary metabolites have valuable functions as antibiotics, toxins or signaling molecules. While many such molecules are produced by filamentous fungi and bacteria, the new research suggests some budding yeasts make secondary metabolites as well.
Another surprising aspect of the study was the finding that many yeast species that do not make pulcherrimin – including S. cerevisiae – nonetheless have working PUL3 and PUL4genes. Patterns across many yeast lineages suggest that retaining these genes allows some species to capitalize on the pulcherrimin made by others, Krause explained.
The findings help highlight the value of stepping beyond traditional lab models.
With a better understanding of the steps involved in pulcherrimin production, the researchers are now poised to try to tweak the production machinery and turn it to making isobutanol instead. “This research is a starting point for taking what we've learned about pulcherrimin and applying it to biofuels,” Krause concluded.
This study was supported by grants from the U.S. Department of Energy (DE-SC0018409 and DE-FC02-07ER6449), the National Science Foundation (DEB-1442148, DEB-1442113, DEB-1253634 and DGE-126259) and the National Institutes of Health (5T32GM007133).

